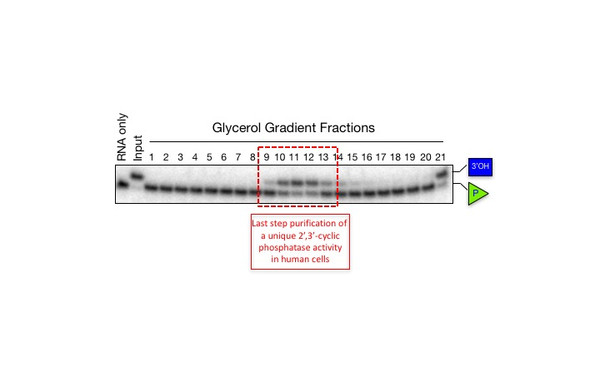
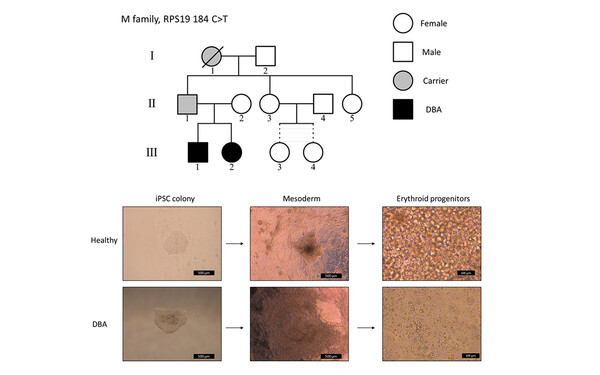
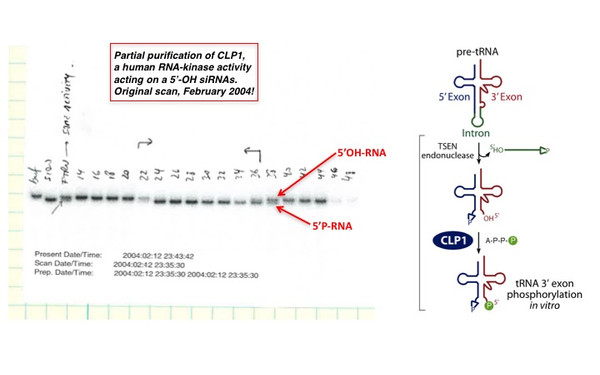
Javier Martinez
Javier Martinez obtained his PhD from the University of Buenos Aires, Argentina. As a Post-Doc at the University of Uppsala, Sweden, he turned to RNA biology and identified and characterize the poly(A) ribonuclease PARN. Later, at the Max Planck Institute in Göttingen and The Rockefeller University in New York, he devoted to RNA interference and purified the RNA-induced silencing complex, RISC. As a Junior Group Leader at IMBA, in Vienna, Javier and his team continued revealing features of RNAi and RISC. However, the discovery of the RNA 5’ kinase CLP1 and the tRNA ligase complex redirected his laboratory towards the enzymatic machineries that catalyze non-canonical RNA splicing, an essential event for the maturation of pre-tRNA molecules and the reshaping of the Xbp1-mRNA during the Unfolded Protein Response. Javier’s lab is currently exploring areas beyond RNA biology. Javier is a Professor of the Medical University of Vienna and his laboratory is located at the Max Perutz Labs, within the vibrant Vienna BioCenter. Javier used to play table tennis and is a football (River Plate, Barcelona and Manchester City) and Formula 1 fan!