The Question
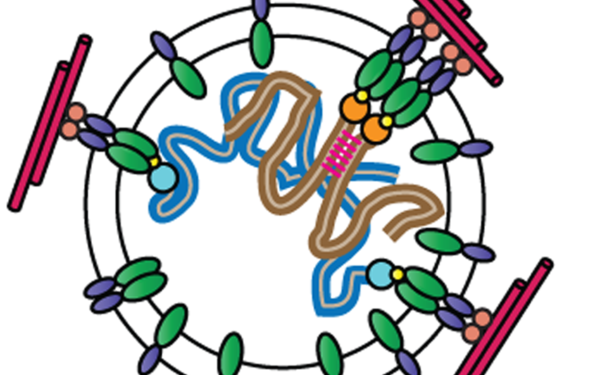
In each generation, the two parental genomes must pair, recombine, and segregate to a newly mixed set of haploid chromosomes in a specialized cell division called meiosis. Failures in this process lead to miscarriages and congenital diseases. Research in my lab is directed towards the identification of genes and processes essential for accurate chromosome segregation during meiosis. For this, we study the meiotic entry network, which ensures the timely coordination and initiation of meiotic processes. We study the mechanisms of recognition and the well-ordered side-by-side alignment of homologous chromosomes. Furthermore, we are interested in the roles of topoisomerases during meiotic recombination, and their function in resolving unwanted chromosomal connections or crossover intermediates. The identification of any new risk factor leading to unfaithful partitioning of chromosomes into gametes is of high relevance to human health.