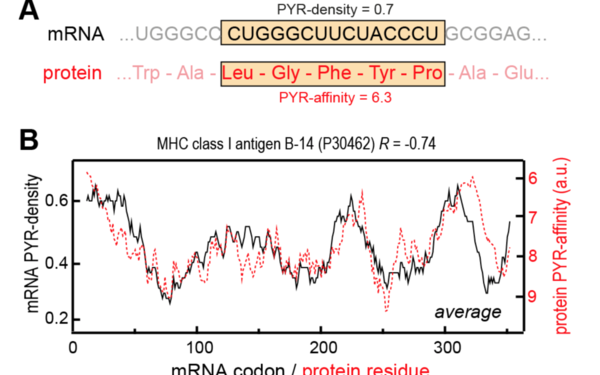
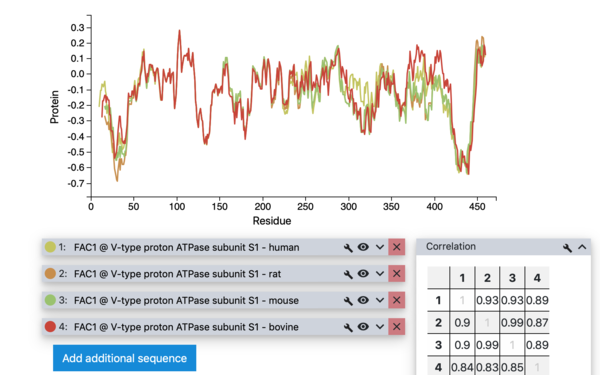
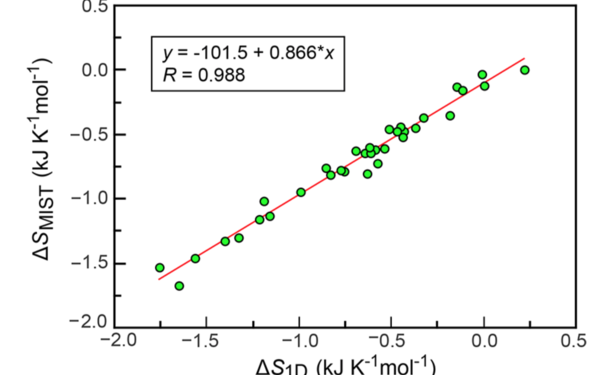
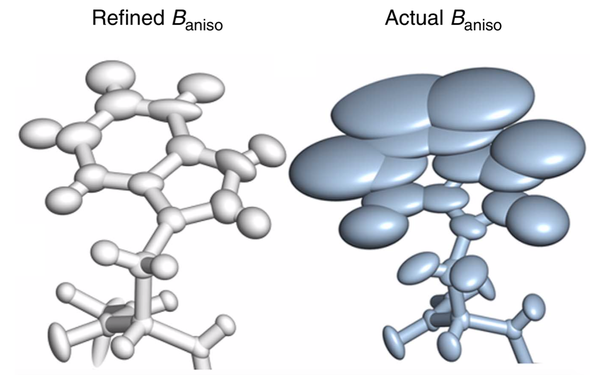
Bojan Zagrovic
Bojan Žagrović obtained an undergraduate degree in biochemistry from Harvard University and a PhD in biophysics from Stanford University, working with Vijay S. Pande. He was an EMBO postdoc with Wilfred F. van Gunsteren at ETH Zurich and a group leader and scientific director at Mediterranean Institute for Life Sciences in native Croatia. He joined Max Perutz Labs and University of Vienna in 2010.