Selected Publications
MINFLUX Achieves Molecular Resolution with Minimal Photons.
2025, Nature Photonics 19, no. 3 (March 2025): 238–47.
DOI: 10.1038/s41566-025-01625-0
Scheiderer, Lukas, Zach Marin, and Jonas Ries.
Skp1 Proteins Are Structural Components of the Synaptonemal Complex in C. Elegans.
2024, Science Advances 10, no. 7 (February 16, 2024): eadl4876.
DOI: 10.1126/sciadv.adl4876
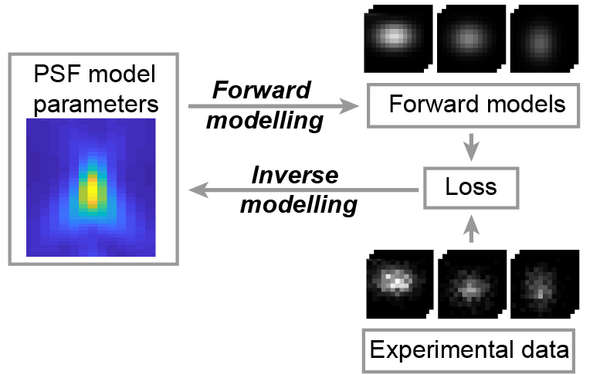
Blundon, Joshua M., Brenda I. Cesar, Jung Woo Bae, Ivana Čavka, Jocelyn Haversat, Jonas Ries, Simone Köhler, and Yumi Kim.
Universal inverse modeling of point spread functions for SMLM localization and microscope characterization.
2024 Nature methods;21(6):1082, 1093, 1082-1093.
PMID: 38831208
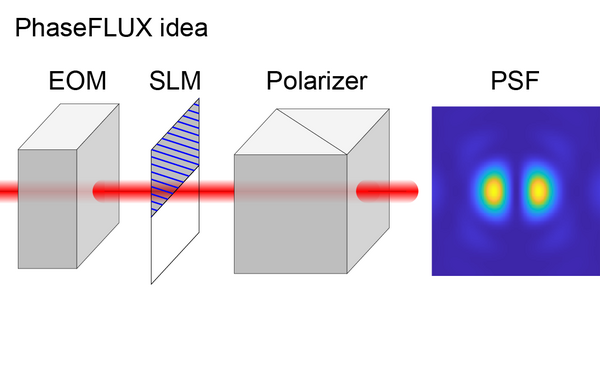
Liu Sheng, Chen Jianwei, Hellgoth Jonas, Müller Lucas-Raphael, Ferdman Boris, Karras Christian, Xiao Dafei, Lidke Keith A, Heintzmann Rainer, Shechtman Yoav, Li Yiming, Ries Jonas
Simple and robust 3D MINFLUX excitation with a variable phase plate.
2024 Light, science & applications;13(1):134.
PMID: 38849346
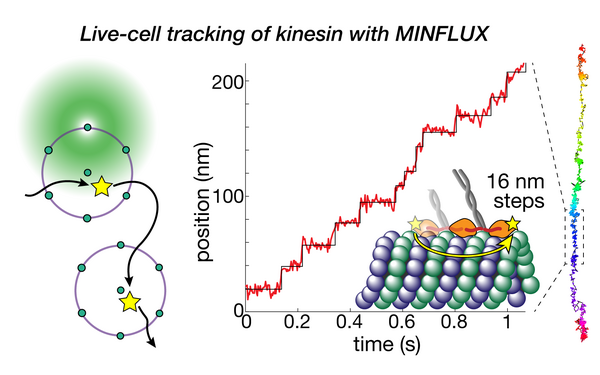
Deguchi Takahiro, Ries Jonas
Direct observation of motor protein stepping in living cells using MINFLUX.
2023 Science (New York, N.Y.)(6636)
PMID: 36893247
Deguchi Takahiro, Iwanski Malina K, Schentarra Eva-Maria, Heidebrecht Christopher, Schmidt Lisa, Heck Jennifer, Weihs Tobias, Schnorrenberg Sebastian, Hoess Philipp, Liu Sheng, Chevyreva Veronika, Noh Kyung-Min, Kapitein Lukas C, Ries Jonas
Maximum-likelihood model fitting for quantitative analysis of SMLM data.
2023 Nature methods(1)
PMID: 36522500
Wu Yu-Le, Hoess Philipp, Tschanz Aline, Matti Ulf, Mund Markus, Ries Jonas
Nanoscale structural organization and stoichiometry of the budding yeast kinetochore.
2023 The Journal of cell biology(4)
PMID: 36705601
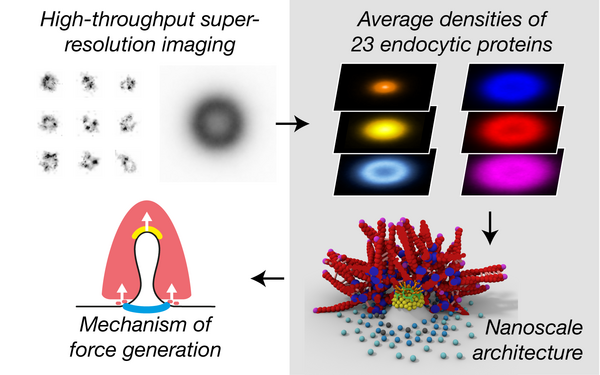
Cieslinski Konstanty, Wu Yu-Le, Nechyporenko Lisa, Hörner Sarah Janice, Conti Duccio, Skruzny Michal, Ries Jonas
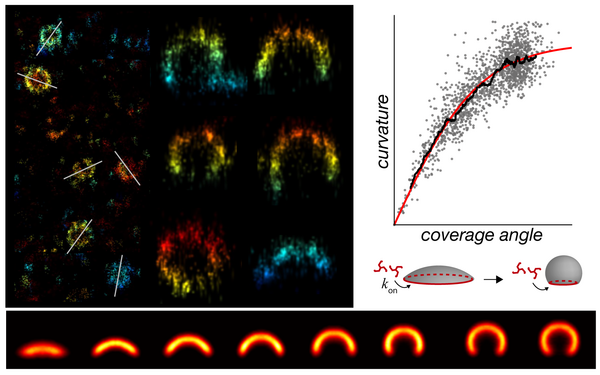
Clathrin coats partially preassemble and subsequently bend during endocytosis.
2023 The Journal of cell biology(3)
PMID: 36734980
Mund Markus, Tschanz Aline, Wu Yu-Le, Frey Felix, Mehl Johanna L, Kaksonen Marko, Avinoam Ori, Schwarz Ulrich S, Ries Jonas
Global fitting for high-accuracy multi-channel single-molecule localization.
2022 Nature communications(1)
PMID: 35668089
Li Yiming, Shi Wei, Liu Sheng, Cavka Ivana, Wu Yu-Le, Matti Ulf, Wu Decheng, Koehler Simone, Ries Jonas
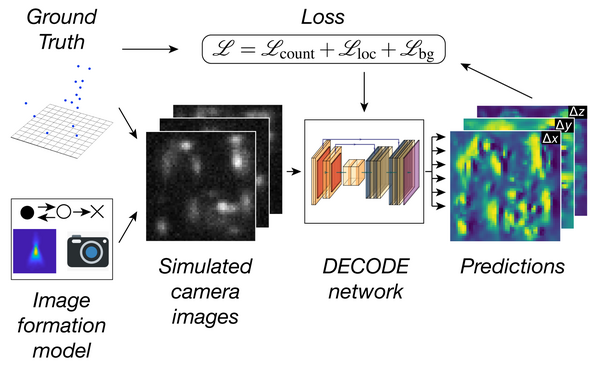
Deep learning enables fast and dense single-molecule localization with high accuracy.
2021 Nature methods(9)
PMID: 34480155
Speiser Artur, Müller Lucas-Raphael, Hoess Philipp, Matti Ulf, Obara Christopher J, Legant Wesley R, Kreshuk Anna, Macke Jakob H, Ries Jonas, Turaga Srinivas C
MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells.
2020 Nature methods(2)
PMID: 31932776
Gwosch Klaus C, Pape Jasmin K, Balzarotti Francisco, Hoess Philipp, Ellenberg Jan, Ries Jonas, Hell Stefan W
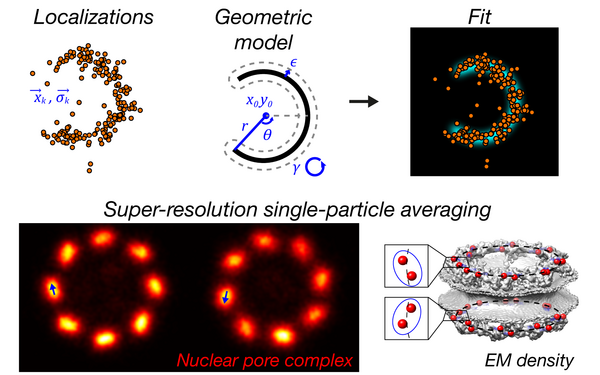
Nuclear pores as versatile reference standards for quantitative superresolution microscopy.
2019 Nature methods(10)
PMID: 31562488
Thevathasan Jervis Vermal, Kahnwald Maurice, Cieśliński Konstanty, Hoess Philipp, Peneti Sudheer Kumar, Reitberger Manuel, Heid Daniel, Kasuba Krishna Chaitanya, Hoerner Sarah Janice, Li Yiming, Wu Yu-Le, Mund Markus, Matti Ulf, Pereira Pedro Matos, Henriques Ricardo, Nijmeijer Bianca, Kueblbeck Moritz, Sabinina Vilma Jimenez, Ellenberg Jan, Ries Jonas
View all Publications of Group Ries