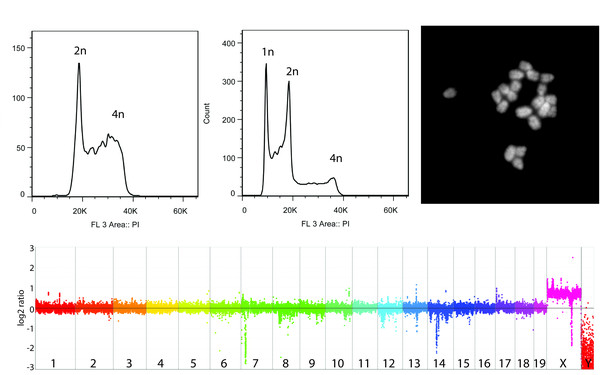
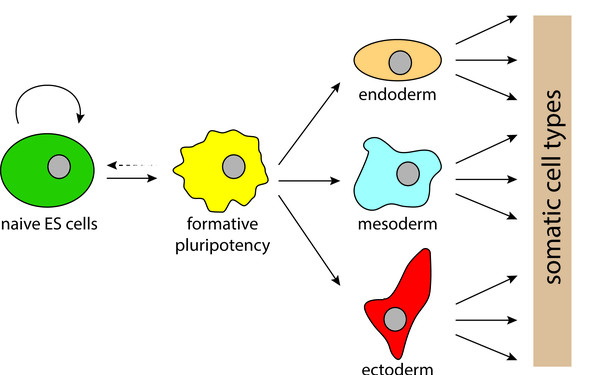
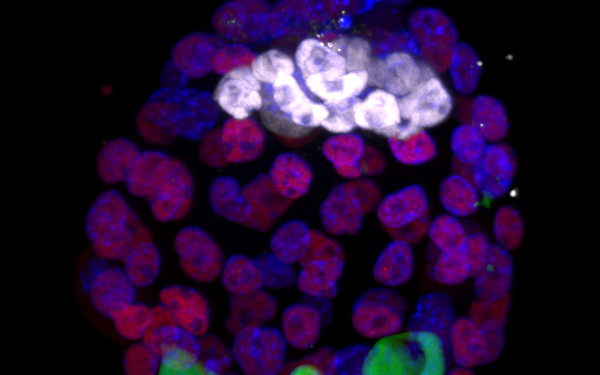
NMD is required for timely cell fate transitions by fine-tuning gene expression and regulating translation.
2022 Genes & development;36(5-6):348, 367, 348-367.
PMID: 35241478
Huth Michelle, Santini Laura, Galimberti Elena, Ramesmayer Julia, Titz-Teixeira Fabian, Sehlke Robert, Oberhuemer Michael, Stummer Sarah, Herzog Veronika, Garmhausen Marius, Romeike Merrit, Chugunova Anastasia, Leesch Friederike, Holcik Laurenz, Weipoltshammer Klara, Lackner Andreas, Schoefer Christian, von Haeseler Arndt, Buecker Christa, Pauli Andrea, Ameres Stefan L, Smith Austin, Beyer Andreas, Leeb Martin
FoxO transcription factors actuate the formative pluripotency specific gene expression programme.
2024 Nature communications;15(1):7879.
PMID: 39251582
Santini Laura, Kowald Saskia, Cerron-Alvan Luis Miguel, Huth Michelle, Fabing Anna Philina, Sestini Giovanni, Rivron Nicolas, Leeb Martin
Genomic imprinting in mouse blastocysts is predominantly associated with H3K27me3.
2021 Nature communications;12(1):3804.
PMID: 34155196
Santini Laura, Halbritter Florian, Titz-Teixeira Fabian, Suzuki Toru, Asami Maki, Ma Xiaoyan, Ramesmayer Julia, Lackner Andreas, Warr Nick, Pauler Florian, Hippenmeyer Simon, Laue Ernest, Farlik Matthias, Bock Christoph, Beyer Andreas, Perry Anthony C F, Leeb Martin
Cooperative genetic networks drive embryonic stem cell transition from naïve to formative pluripotency.
2021 The EMBO journal;40(8):e105776.
PMID: 33687089
Lackner Andreas, Sehlke Robert, Garmhausen Marius, Giuseppe Stirparo Giuliano, Huth Michelle, Titz-Teixeira Fabian, van der Lelij Petra, Ramesmayer Julia, Thomas Henry F, Ralser Meryem, Santini Laura, Galimberti Elena, Sarov Mihail, Stewart A Francis, Smith Austin, Beyer Andreas, Leeb Martin
Genetic exploration of the exit from self-renewal using haploid embryonic stem cells.
2014 Cell stem cell(3)
PMID: 24412312
Leeb Martin, Dietmann Sabine, Paramor Maike, Niwa Hitoshi, Smith Austin
Derivation of haploid embryonic stem cells from mouse embryos.
2011 Nature(7371)
PMID: 21900896
Leeb Martin, Wutz Anton
Polycomb complexes act redundantly to repress genomic repeats and genes.
2010 Genes & development(3)
PMID: 20123906
Leeb Martin, Pasini Diego, Novatchkova Maria, Jaritz Markus, Helin Kristian, Wutz Anton