
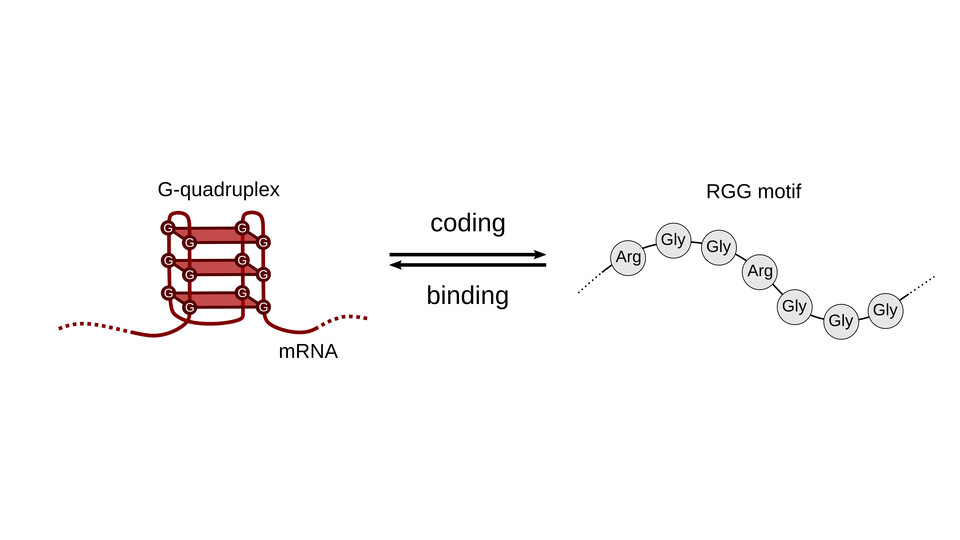
RNA G-quadruplexes (rG4s) are three-dimensional secondary RNA structures formed by guanine residues that are thought to play a role in RNA-protein interactions. Intriguingly, the RGG motif, an amino acid sequence rich in arginine (R) and glycine (G) and known to interact with rG4s, is encoded by mRNA sequences that are guanine-rich, establishing a link between rG4-containing RNAs and proteins with RGG motifs. Using a computational analysis, the Žagrović lab now shows that hundreds of RGG motifs in human are indeed encoded by rG4s in their mRNAs.
By analyzing hundreds of RGG motifs in the human proteome and examining G-quadruplexes in their corresponding mRNAs, the researchers have discovered that especially long RGG motifs are likely to be associated with rG4s in their mRNA. First author Marlene Adlhart explains: “Our study is the first to demonstrate that a peptide motif and an RNA secondary structure are not only connected physically and interact via direct binding, but that their interaction may reflect the structure of the genetic code.”
Perutz group leader Bojan Žagrović explains further: “Our study addresses the intriguing idea that mRNA and the proteins they encode exhibit physical complementarity and bind, despite their significant size difference, with mRNA being five times longer than its protein counterpart.” To bridge this gap, the lab suggests that RNA adopts compact structures, such as G-quadruplexes, effectively shortening the mRNA and, thereby, facilitating its interaction with the protein it encodes.
A significant question about many RNA-binding proteins – such as FMRP, FUS, and G3BP1, which are known for their long, repetitive RGG motifs and their ability to bind rG4s in target mRNAs – has also been addressed: What could be the mechanism these proteins use to recognize their own mRNAs that encode them? Bojan explains: “Our study suggests that RGG-rG4 interaction, which is hardwired in the structure of the genetic code, may facilitate the establishment of autoregulatory feedback loops in the cell.”
Highlighting the broader implications of the study, he adds: “Our findings open exciting new directions, including investigating other RNA-binding motifs like RNA recognition motifs (RRMs) or zinc fingers, which might similarly prefer to bind the mRNAs that encode them. This could significantly advance our understanding of RNA-protein interactions.”
DOI: 10.1073/pnas.2413721122