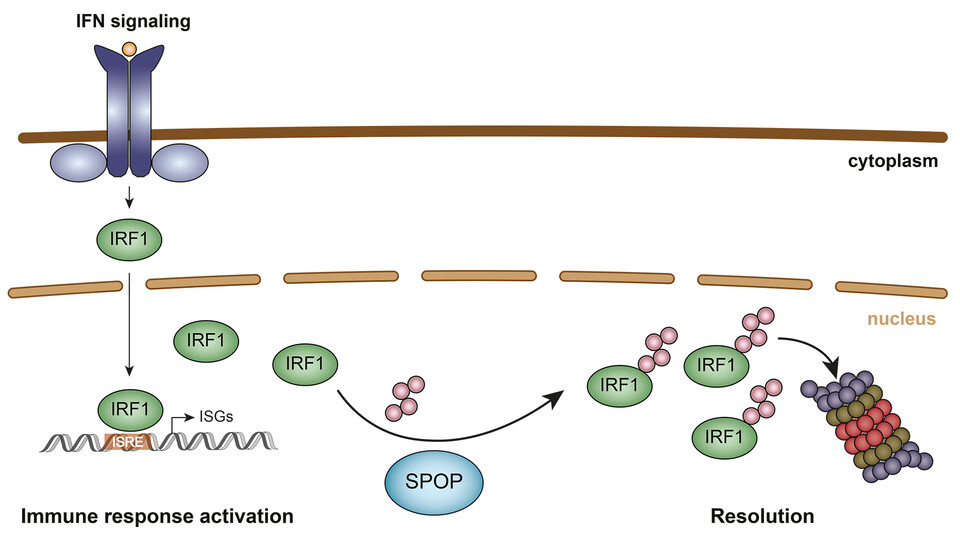


Cell survival depends acutely on robust responses to environmental insults, including invading pathogens such as bacteria and viruses. Signaling pathways that sense the presence of the pathogen upregulate key transcription factors that subsequently drive the expression of response genes. These response genes include secreted proteins called interferons that instruct neighboring cells to mount an immune response to counter the threat. Interferon Regulatory Factor 1 (IRF1) is key to this response, but too much of it triggers hyperactivation of the pathway and is associated with a range of inflammatory diseases. IRF1 levels are controlled both transcriptionally and at the protein level, but precisely how IRF1 is targeted for degradation is not clear.
Using a genome-wide CRISPR knockout screen, the Versteeg lab sought to identify regulators of IRF1 abundance in an unbiased manner. Knocking out the Cullin E3 ubiquitin ligase adaptor Speckle Type BTB/POZ Protein (SPOP) elevated IRF1 levels by 40% across different cell lines. E3 ubiquitin ligases are enzymes that covalently couple ubiquitin to proteins that are targeted for degradation. Ubiquitin is recognized by the proteosome, which unfolds and degrades the ubiquitinated substrate. The scientists showed that SPOP and IRF1 co-localize in the nucleus, an essential pre-requisite for SPOP-mediated proteosomal degradation.
The specificity of protein degradation is conferred by sequence-specific ‘degrons’ within the target. SPOP interaction with IRF1 depends on the SPOP MATH domain and serine/threonine-rich degrons in IRF1. To investigate the relevance of SPOP for IRF1-stimulated gene transcription programs, the scientists examined the effect of SPOP co-expression on IRF1-driven luciferase expression in cells. While wild-type SPOP reduced luciferase expression by an order of magnitude, mutation of its MATH domain or the degrons in IRF1 failed to elicit an effect.
The anti-viral state elicited by IRF1 is dependent on its expression levels. Consistently, exogenous over-expression of IRF1 reduced virus titers in Vesicular Stomatitis Virus (VSV)-infected cells. When the scientists also knocked out SPOP, the virus titers were reduced by a further 10-fold, indicating that SPOP is a negative regulator of IRF1 and an important brake on the innate immune response. SPOP joins a growing list of proteins that regulate IRF1 in a variety of cellular contexts.
The work was a team effort involving collaborators from the Zuber lab (IMP - Institute for Molecular Pathology). Co-first authors Irene Schwartz, Milica Vunjak and Valentina Budroni were instrumental in developing the story. Irene Schwartz says: “This study is an example of how genetic screens can be combined with biochemistry and cell biology to uncover new biology. Without my teammates and our collaborators, this work would not have been possible.”