The Mass Spectrometry Facility provides proteomics services primarily to the research groups of the Max Perutz Labs Vienna and if capacity remains also to external customers. We apply state-of-the-art LC-MS analysis platforms and bioinformatics tools for identifying and quantifying peptides and proteins, including their post-translational modifications. In addition, we offer consultation, training, and capacities for method development to our users to help finding the best analytical solution for a given problem.
Our services comprise the following applications:
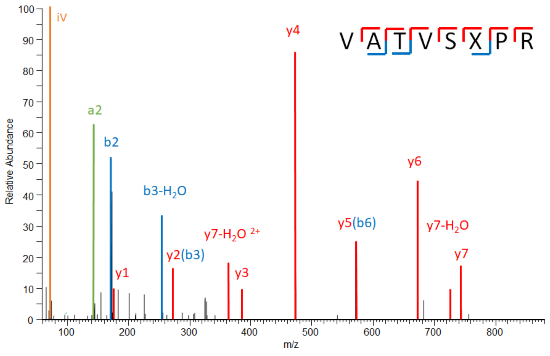
Identification
This includes protein digestion (from SDS-gel, in-solution, or beads), reduction and alkylation, a quality control run on an LC-UV-VIS system, high resolution accurate mass LC-MS/MS analysis, database search, qualitative data analysis, and the generation of a report file. We perform analysis of post-translational modifications with or without prior enrichment. We have extensive experience in the analysis of phosphorylation and lysine acetylation but we are open to integrate any other PTM of interest, if it is accessible by means of MS.
For more information concerning sample submission and sample preparation please have a look at our guidelines here.
Quantification
Our pipelines support quantitation of peptides and proteins, using metabolic (SILAC) or chemical (e.g. TMT, iTRAQ, dimethyl) labels as well as label-free approaches. Properly applied, these methods allow reliable quantification of a large number of proteins and their (modified) peptides in complex samples. However, careful experimental planning is key to the generation of useful data. In case you would like to perform such experiments we would be happy to assist in the planning phase.
For more information concerning sample submission and sample preparation please have a look at our guidelines here.
Interaction proteomics
Protein affinity enrichment followed by mass spectrometry has become a major technology for determining protein interactors. In our workflow we apply label-free quantification to distinguish between background proteins and true interactors. If performed in sufficient replicates this allows comparative statistical analysis to define interaction partners with higher confidence.
For more information concerning sample submission and sample preparation please have a look at our guidelines here.
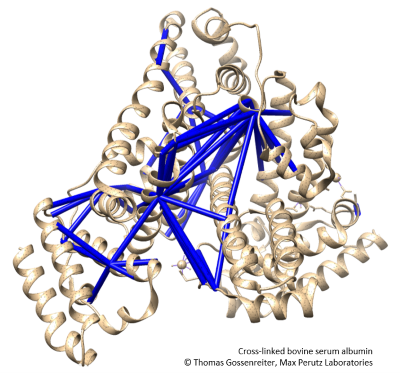
Cross-linking mass spectrometry
Cross-linking mass spectrometry has evolved into a powerful technique which provides structural information and modelling constraints on proteins and protein complexes in solution. We offer a specialised workflow for your cross-linked protein samples including digestion in gel or in solution, LC-MS/MS measurement, and data analysis.
For more information concerning sample submission and general sample preparation please have a look at our guidelines here.
For cross-linking specific guidelines please follow the link here.
Please note that this service is only available to groups from Max Perutz Labs, University of Vienna, and Medical University of Vienna.
Intact protein mass analysis
Determining the mass of an intact protein as accurately as possible can serve as quality control of expressed proteins, help to characterize possible chemical or post-translational modfications, examine truncation products, etc. We provide intact protein mass analysis under denaturing conditions using a high-resolution and accurate mass ESI-TOF-MS system, coupled to a micro-flow HPLC system for online desalting and separation. Native protein (complex) analysis and ion-mobility separation are not offered as a routine service.
For more information concerning sample submission and sample preparation please have a look at our intact protein MS guidelines here.
Please contact the team before submission of samples.
Sample drop-off is at our facility, which is located at VBC6, Dr.-Bohrgasse 7. Please ask at the entrance to give us a call and we will come to meet you there. We are available between 9 am to 5 pm but please give us a heads-up before to make sure somebody is available in the office.
For information on internal fees and for submission forms please visit the intranet:
Please contact the team before submission of samples.
Sample drop-off is at our facility, which is located at VBC6, Dr.-Bohrgasse 7, 1030 Vienna. Please ask at the entrance to give us a call and we will come to meet you there. We are available between 9 am to 5 pm but please give us a heads-up before to make sure somebody is available in the office.
Office:
VBC6
Dr. Bohr Gasse 7, 1030 Vienna
Office: room E72
Internal users of the Max Perutz Labs: please visit the intranet for current rates.
External users: Please contact us for a quote tailored to your specific needs. Please bear in mind that internal users have priority and external service can only be offered when there are free capacities.
All services and data are provided on a best effort basis “as is” and for research purpose only. The Max Perutz Labs make no warranties of any kind with respect to performance of any services and quality of provided data, including warranties of merchantability or fitness for any particular purposes of such data, material or information. We will treat your samples with utmost care and apply the most sensible procedures existing to our knowledge to provide you with the information that you need. Despite these efforts we cannot guarantee that all analyses will turn out to be successful, due to the intrinsic limitations of the sample preparation process and liquid chromatography/mass spectrometry. This method-specific risk lies with the customer and, as a consequence, we will charge every run, irrespective of whether a desired result was obtained. In cases when technical or processing problems occur in the facility we will not charge for this sample or run. Please keep in mind that the analytical question may require specific measurement parameters, which need to be adjusted before the measurement. As a consequence it is important to specify the exact aim of the measurement and all relevant details before sample processing. We reserve the right not to measure particular samples if their quality does not meet our criteria (as determined in an LC-UV quality control run) and threatens to damage the instrumentation. All costs for sample handling before quality control will be invoiced to the customer.
We have shared access to the following instruments which are owned and maintained by Vienna Biocenter Core Facilities (VBCF):
Thermo Exploris 480
Thermo Eclipse
Bruker timsTOF HT
Waters Synapt G2-Si HDMS
Most of the mass spectrometers are coupled to Thermo Scientific Vanquish Neo systems for reliable, high performance separation of the samples. For more detailed information on the different instruments please visit the website of the VBCF.
Based on the application and the research question we apply different software packages and search engines. Currently these include FragPipe, MaxQuant, Spectronaut, pLink, Merox, and Skyline. For downstream statistical analysis we use Perseus, Python and R. All scripts developed in house and custom scripts for publications are released on our GitHub-repository.
Acknowledgements or co-authorships are very important for core facilities as they facilitate raising funds and support in the future. For this reason we request acknowledgement for the use of the facility services in your publications according to the following logic:
For routine analysis we kindly ask you to mention the facility (as "Max Perutz Labs Mass Spectrometry Facility") in the acknowledgement section of your manuscript.
A suggested minimum format would be:
“Proteomics analyses were performed by the Mass Spectrometry Facility at Max Perutz Labs using the VBCF instrument pool.”
If a service was provided through the Vienna Life Science Instruments Initiative (VLSI) please add “a member of VLSI” after “Max Perutz Labs”.
If a facility team member develops novel resources or methods, contributes substantially to the experimental design, or performs advanced data analyses which are beyond the invoiced routine service and which are an important part of the publication a co-authorship seems appropriate.
This policy is also suggested by the Association of Biomolecular Research Facilities (ABRF) which has published a guideline on this matter. If you think that this could become an issue just let us know as early as possible, ideally before the project starts, and we will try to find a solution.