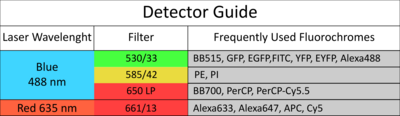
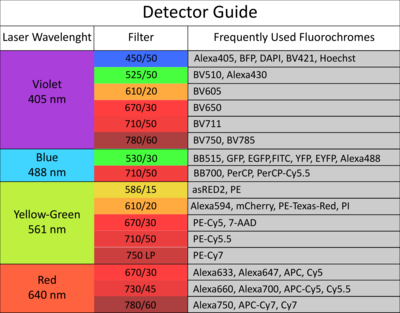
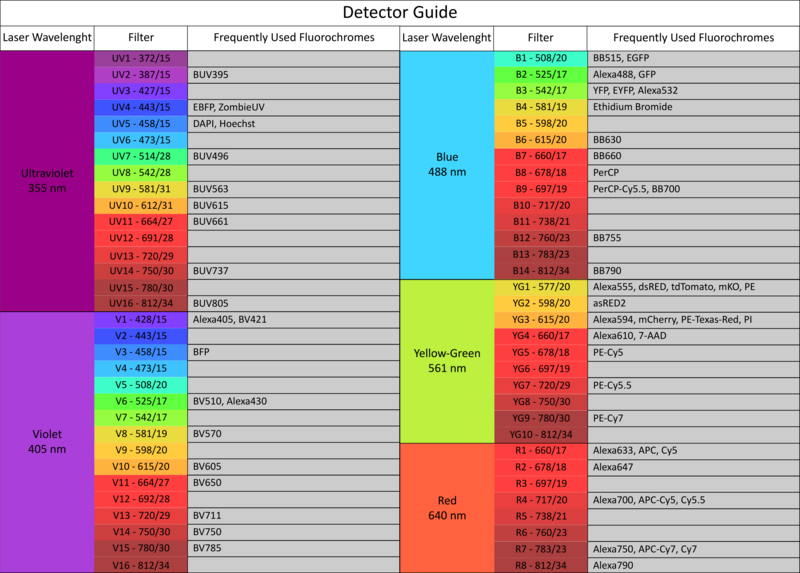
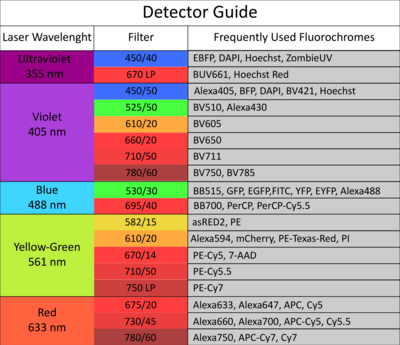
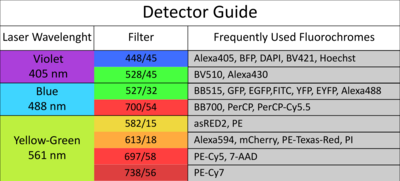
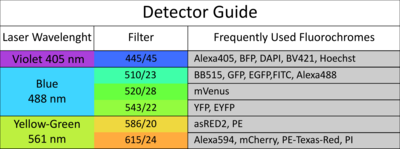
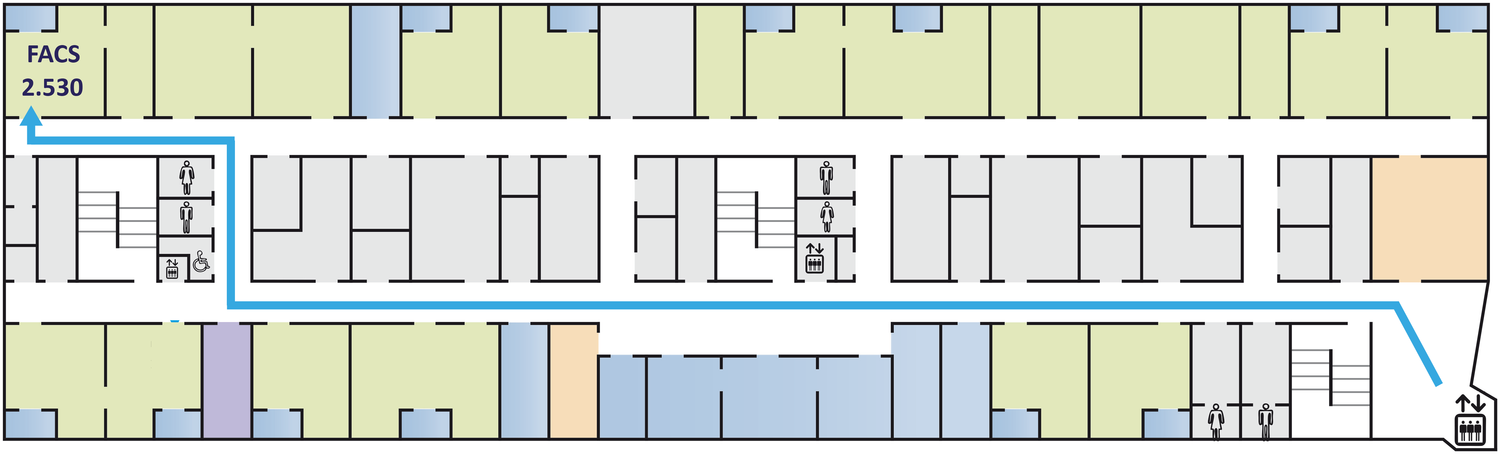
The facility offers access to state-of-the-art cell and large particle sorting technologies and analysis tools, while also providing dedicated support and adaptable configurations to assist researchers.
The facility offers access to state-of-the-art cell and large particle sorting technologies and analysis tools, while also providing dedicated support and adaptable configurations to assist researchers.
We are partner of the VLSI Programm (Vienna Life-Science Instruments). https://www.vlsi.at/