
The great Dutch microscopist Antonie van Leeuwenhoek first described ciliated organisms of the gut microbiome in the late 1600s. Since then, cilia have been observed on the surface of eukaryotic cells from green algae to humans, and their structures and functions described in ever-increasing detail. Cilia not only have a long history of study, but are also evolutionarily highly ancient, with their emergence thought to date back to the last common ancestor of all eukaryotes over 1 billion years ago. However, despite being ancient organelles, cilia are not universally conserved. Accumulating evidence of their biomedical importance has led to considerable efforts over the past decades to define the set of genes required for their formation and function. Evidence, however, suggests that our current list of genes is as yet incomplete, with the causative mutation in many ciliopathy patients still unknown and key steps in cilium assembly remaining poorly understood.
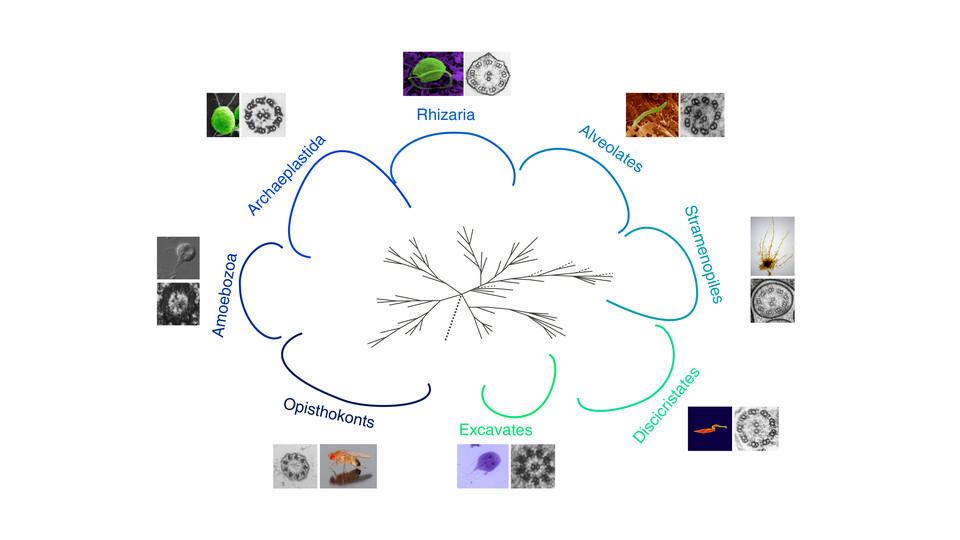
Armed with the recent explosion in the number of fully sequenced eukaryotic genomes across all major phyla and the observation that non-ciliated organisms rapidly lost cilia-associated genes, the Dammermann lab sought to identify ciliogenesis genes across all eukaryotes. Using simple patterns of co-occurrence across evolution to predict genes with common functions, the lab identified a set of 386 genes associated with the key steps of centriole assembly, cilium biogenesis and motility. Of these, more than a third (152) were previously uncharacterized or had not been associated with cilium biogenesis or function.
Using the nematode worm, Caenorhabditis elegans, and the fruit fly, Drosophila melanogaster, as model organisms, the researchers experimentally validated their findings by systematically depleting each candidate gene. The team found that most, if not all, of the novel genes are likely cilium-associated. Using a wide array of microscopy-based phenotypic profiling assays, the scientists were able to identify and further characterize a set of novel genes involved in the as yet poorly understood steps of early cilium biogenesis.
The compendium of genes identified in this study is remarkable for two reasons: first, all 386 genes can be traced back to the last eukaryotic common ancestor. This implies an astonishing degree of cellular complexity of the ancestral eukaryote. Secondly, only 15 genes are universally conserved in ciliated species and, therefore, indispensable for formation of the cilium. As project leader Alex Dammermann says, "in this case, many roads apparently lead to Rome." The addition of 152 novel genes opens new frontiers in basic research into ciliogenesis and may lead to advances in our understanding of human ciliopathies.
Publication:
Jeroen Dobbelaere, Tiffany Y Su, Balazs Erdi, Alexander Schleiffer, Alexander Dammermann: A phylogenetic profiling approach identifies novel ciliogenesis genes in Drosophila and C. elegans. The EMBO Journal (2023).