
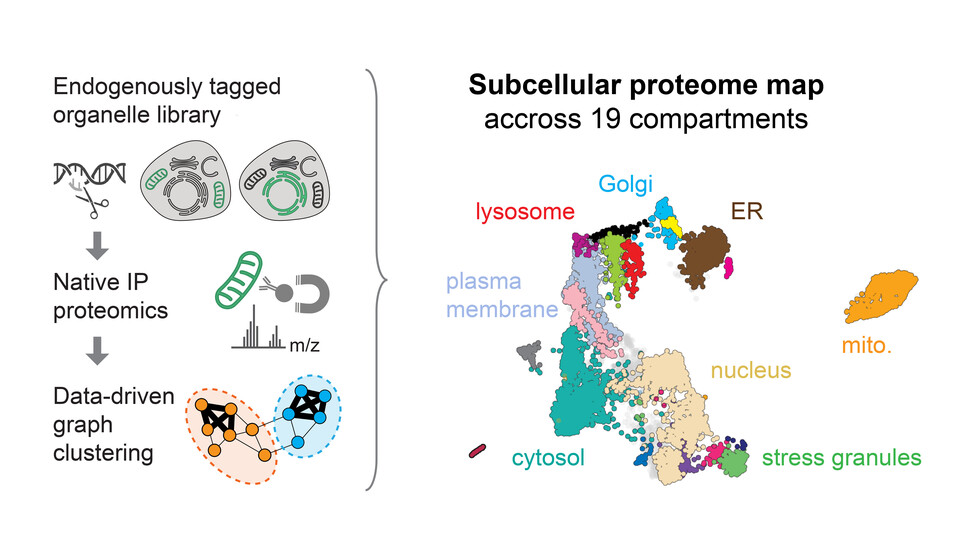
Organelles are a defining feature of eukaryotic cells, organizing molecules and processes in specific compartments within the cell. While fluorescence microscopy, subcellular fractionation, and proximity labeling coupled to mass spectrometry have been very successful in determining the distribution of individual proteins within these compartments, the precise localization of many proteins remains unknown due to limitations in resolution and throughput. To address these challenges, Marco Hein and the team at the Biohub developed a novel approach for the comprehensive subcellular characterization of the human proteome.
The researchers combined subcellular fractionation with targeted enrichment of various organelles using specific marker proteins, followed by mass spectrometry to quantify the associated proteomes. This approach yields a precise, static snapshot of protein distribution within the cell. Perutz group leader Marco Hein explains: “Our protocol aims to break down the cell mechanically while largely preserving the structural integrity of subcellular structures. This method allows us to isolate all organic material linked to the marker protein.” Using markers for 36 subcellular compartments, the researchers improved the resolution and consistency of their data.
In the second part of the study, the authors sought to understand how cells adjust their proteomes in response to viral infection. By infecting cells with the coronavirus OC43, a close relative of SARS-CoV-2, the researchers modeled the dynamic re-localization of proteins. While most proteins were unaffected by viral infection, many of those that did change locations were linked to ferroptosis, an iron-dependent, regulated cell death pathway. “We were surprised to see that protein re-localization revealed a set of proteins that shows minimal overlap with those proteins that are regulated at the level of gene expression or absolute protein abundance. Profiling the remodeling of subcellular organization gave us unique insights into the pathways regulating infection that we would not have discovered otherwise”, says project leader Manuel Leonetti from the Biohub.
“I am confident that our subcellular map will become a go-to resource for all cell biologists who want to know where their proteins of interest are localized within the cell”, says Marco Hein. “In addition, our work offers a framework for future studies, supporting scientists in the exploration of protein behavior in response to cellular perturbations.”
DOI: 10.1016/j.cell.2024.11.028